Type 1 diabetes (T1D) and celiac disease (CD) are the two most common autoimmune childhood diseases that share their HLA-DQ2 and DQ8 genetic origin. There has been a current increase in both diseases worldwide. In children from the low-population State of Sonora (15 inhabitants/km2) in northwestern Mexico, there is no information on their genetic risk or the distribution of the related alleles in the general population.
AimsTo compare the HLA-DQ allele frequency in a representative sample of newborns from Sonora with that of T1D and CD patients to determine the risk gradient and identify the presence of celiac autoimmunity in the T1D group.
MethodsThe study included 397 Sonoran newborns, 44 cases of T1D, and 25 CD cases. The CD and T1D cases were clinically diagnosed by specialists at the Hospital Infantil del Estado de Sonora and the autoantibodies were determined by ELISA. Whole blood was collected, gDNA was extracted, and HLA-DQ2 and DQ8 were typed by PCR-SSP. The risk gradient was calculated by comparing the allele frequencies of the cases with those of the newborns.
ResultsThe Sonoran HLA-DQ risk heterodimer proportion was 16.1% for HLA-DQ2 and 13.6% for HLA-DQ8 with an HLA-DQ2: HLA-DQ8 ratio of 1.2:1. The DQ8/DQ2 genotype represented a 1:14 risk for T1D, whereas the DQ8/DQB1*0201 combination resulted in a 1:6 risk for CD. The prevalence of CD autoimmunity in T1D children was 7%.
ConclusionThe Sonoran population has a distinctive HLA-DQ allele distribution due to its ancestry. The HLA-DQ8 combinations with DQ2 or one of its alleles conferred the highest risk for both diseases and T1D and CD frequently appear together.
La diabetes tipo 1 (DT1) y la enfermedad celíaca (EC) son 2 enfermedades autoinmunes frecuentes en la infancia y comparten su predisposición genética (HLA-DQ2 y DQ8). La prevalencia de ambas se ha incrementado en el mundo. En el estado de Sonora (15 habitantes/km2), se desconoce información sobre su riesgo genético o la distribución de los alelos asociados en la población general.
ObjetivoComparar la frecuencia alélica HLA-DQ de una muestra representativa de recién nacidos sonorenses con la de pacientes DT1 y EC para determinar el gradiente de riesgo e identificar la presencia de autoinmunidad celíaca en el grupo de DT1.
MétodosSe incluyeron 397 recién nacidos sonorenses, 44 casos DT1 y 25 EC, diagnosticados clínicamente y con autoanticuerpos por ELISA. Se colectó una muestra de sangre, se extrajo ADNg y tipificaron HLA-DQ2 y DQ8 por PCR. El gradiente de riesgo se calculó comparando las frecuencias alélicas de los casos con respecto a los recién nacidos.
ResultadosLa proporción de heterodímeros de riesgo en sonorenses fue de 16.1% para HLA-DQ2 y de 13.6% para HLA-DQ8 con una proporción HLA-DQ2:DQ8 de 1.2:1. El genotipo DQ8/DQ2 representó un riesgo de 1:14 para DT1, mientras que para EC el DQ8/DQB1*0201 generó un riesgo de 1:6. La prevalencia de autoinmunidad asociada a EC fue del 7% en los niños DT1.
ConclusiónLos sonorenses tienen una distribución de alelos HLA-DQ distintiva debido a su ascendencia. Las combinaciones del HLA-DQ8 con DQ2 o uno de sus alelos confirieron el máximo riesgo para ambas enfermedades. La DT1 y EC frecuentemente se presentan juntas.
Type 1 diabetes (T1D) is a chronic disease caused by autoimmune-mediated destruction of pancreatic beta cells, usually leading to absolute insulin deficiency1. Celiac disease (CD) is another inflammatory systemic disease, primarily affecting the small intestine, caused by an immune-based reaction to dietary gluten (storage protein for wheat, barley, and rye) in individuals with a genetic predisposition and it is resolved by excluding gluten from the diet2.
T1D and CD are the 2 most common autoimmune childhood diseases, with a prevalence of 1:100 to 1:200 for CD3 and 1:300 for T1D4. In addition, the prevalence of CD in T1D patients appears to be 20 times higher5, being frequently under-diagnosed because adherence to diagnostic guidelines in current practice varies widely6. These immune-mediated diseases share some characteristics involving environment and genetics7.
The class II human leukocyte antigen (HLA) is the single most important genetic factor for autoimmune diseases such as T1D and CD, mainly the DQ and DR isotypes8. Thus, the HLA DQ2 and DQ8 haplotypes contribute to over 50% of the genetic load for both disorders9,10. Expressed HLA-DQ molecules are heterodimers of α and β polymorphic chains that can be encoded in cis or trans11. In antigen-presenting cells of the immune system, these molecules specifically bind peptides, such as those derived from gluten that induce CD, as well as peptides from islet autoantigens, including insulin, that degenerate in T1D12.
In Europe, around 90-95% of celiac patients have the HLA-DQ2 haplotype, composed of the DQA1*0501/DQB1*0201 alleles, while the other 5-10% have the HLA-DQ8 haplotype with DQA1*0301/DQB1*030213. These same haplotypes are present in about 90% of T1D patients14. However, a higher risk has been associated with HLA-DQ8, in contrast to CD, in which HLA-DQ2 provides the greatest risk15. Although other factors have been studied for these diseases, such as affected relatives, dietary pattern, and early intestinal infections, the HLA risk genotype is the only known factor that is significantly associated with the development of CD autoimmunity16. Therefore, the possibility of stratifying the individuals based on their HLA-DQ status is necessary to implement preventive and therapeutic targeted interventions for these diseases17,18. This approach would favor personalized health care development, adapted to the local characteristics of each population.
In northwestern Mexico, the State of Sonora presents unique characteristics with respect to its territory and inhabitants. Sonora is the second largest state in Mexico, with an area of 179,516km2 and a population of 2,662,480 people19, corresponding to a population density of less than 15 inhabitants/km2. In addition, due to the fact that most of the Sonoran territory is desert, inhabitants have remained relatively isolated from the rest of the country. Thus, the Sonoran admixture population has the highest European and the lowest Amerindian ancestry contribution compared with other Mexican populations20. However, no data on the frequency of the HLA-DQ2 and DQ8 alleles in the general population are known to date.
There has been a current increase in cases of T1D and CD in Sonoran children whose population is a special admixture group. Therefore, the aim of this study was to compare the HLA-DQ allele frequency in newborns from Sonora with patients that have T1D and CD to determine their risk gradient and to identify the presence of celiac autoimmunity in the T1D group.
MethodsSubjectsAn analytical cross-sectional study was conducted in Sonora, Mexico, with children seen at the Hospital Infantil del Estado de Sonora. It is the largest regional children's hospital and at least 90% of the parents and grandparents of the patients were born in the Mexican Northwest 21.
The appropriate sample size for the reference group was determined by the formula for proportions, n=(Z2αpq)/d2, taking into account the total Sonoran population (2,662,384 people), an estimated 30% prevalence of HLA DQ2 and DQ8 haplotypes in other populations, and a confidence level of 95% with a margin of error of 5%. Similarly, for the T1D and CD case groups, the expected prevalence of 1:300 and 1:200 was considered, as well as the fact that more than 90% of the patients with these diseases have the DQ haplotypes. In accordance with this approach, 323 controls, 16 CD cases, and 8 T1D cases were required.
Based on the above, the control sample consisted of 397 infants, born at the end of 2010 and the beginning of 2011 and accounting for 14.5% of the newborn babies at the Hospital Infantil del Estado de Sonora during the collecting period. For the case groups, a total of 44 children and adolescents with T1D and 25 with CD, ranging from 2 to 18 years of age, were recruited during the years of 2012-2014. The patient sample represents the totality of T1D and CD cases detected in that hospital during the sampling period. CD diagnoses were confirmed by the Marsh score determined through small bowel biopsy and anti-tissue transglutaminase (tTG)/anti-gliadin serum antibodies. Likewise, T1D diagnoses were established according to the American Diabetes Association criteria1, including positive anti-GAD and/or anti-IA-2 antibodies.
Ethical approvalAll procedures performed were approved by the Ethics Committee of the Center for Food Research and Development (CIAD) and by the Hospital Infantil del Estado de Sonora Learning and Research Board, in accordance with the standards of the 1964 Helsinki Declaration and its Good Clinical Practice guidelines. Informed consent was obtained from the legal guardians of all individual participants included in the study.
Sample collectionUmbilical cord blood was collected at the time of delivery in all the controls and stored at 4°C for HLA typing. Furthermore, 5mL of peripheral blood of all T1D and CD cases were collected, from which 2mL were taken for DNA extraction. The serum from the remaining sample was then separated and reserved for serologic screening.
DNA extraction and HLA typingGenomic DNA was extracted from whole blood samples using the QIAamp DNA Mini Kit (Qiagen®, Hilden, Germany), according to the manufacturer's instructions. HLA DQ2 and DQ8 typing was performed using the polymerase chain reaction with sequence specific primers (PCR-SSP) for the DQA1*0501, DQA1*0301, DQB1*0201, and DQB1*0302 alleles (IDT-Integrated DNA Technologies, Tucson, AZ, USA), as reported by Olerup et al.22 The amplified PCR products obtained were analyzed on 1.8% agarose gels, stained with GelRed™ (Biotium Inc., Hayward, CA, USA), and visualized under ultraviolet light (Molecular Imager®Bio-Rad, Hercules, CA, USA). The HLA DQ2 and DQ8 haplotypes were confirmed assessing the DRB1*03 and DRB1*04 associated alleles from the HLA-DR fraction through PCR-SSP.
The HLA allele frequencies in our sample were calculated by the direct counting method and compared through a Chi-squared test using NCSS analysis software version 2007. A p value < 0.05 was considered statistically significant.
Risk gradientAfter the 4 HLA-DQ risk allele assessment, 16 possible positivity patterns were expected, thus obtaining the different combinations in which the HLA-DQ risk alleles were present in both the T1D and CD cases. These combinations were used for the analysis. For nomenclature purposes, the term DQ2 was used only when a subject carried the DQA1*0501 allele and the DQB1*0201 allele. Likewise, DQ8 referred only to subjects with both the DQA1*0301 and the DQB1*0302 allele. The subjects’ phenotypes encoded by only one of these alleles, forming dimers with a different chain, were named A1*0501, B1*0201, A1*0301, or B1*0302, respectively.
T1D and CD risks were expressed as 1:N, where N is the number of individuals among whom one patient is present, as previously described by Megiorni et al.23 Thereby, N was calculated as the percentage of the controls (Sonoran newborns from the general population) with that particular HLA-DQ status multiplied by 100 and divided by the percentage of patients with the same DQ typing. For this analysis, we considered the reported prevalence of 1:300 for T1D and 1:100 to 1:200 for CD, placing the results into perspective. To confirm the strength of the estimated gradient, odds ratios (OR) were calculated for each combination of alleles.
Serologic ScreeningThe T1D diagnoses were complemented by anti-GAD and anti-IA-2 antibody quantification with a commercial ELISA kit according to the company's instructions (Kronus®, USA). Anti-transglutaminase IgA and anti-gliadin IgG and IgA titers from serum were also measured by ELISA, using enzymatically digested gliadins extracted in our laboratory. ELISA was run as previously performed in our lab24, including the definition for the cutoff values.
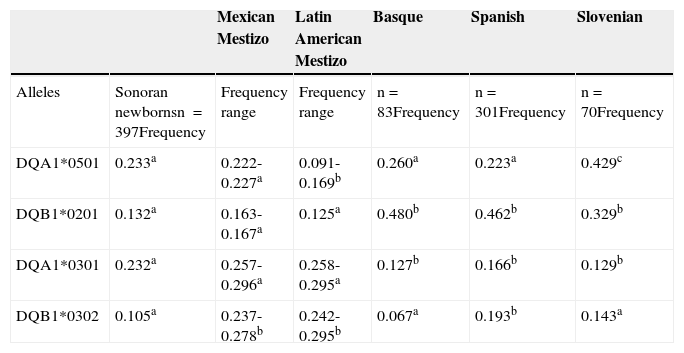
ResultsThe risk heterodimer frequency proportion for newborns from Sonora in northwestern Mexico was 16.1% for HLA-DQ2 and 13.6% for HLA-DQ8, with a HLA-DQ2:HLA-DQ8 ratio of 1.2:1. The HLA-DQ2.5 and HLA-DQ8 allele frequencies in newborns are presented in Table 1. The third and fourth columns show the range of allele frequencies from other populations, such as Mexican Mestizos25–27 and Latin American Mestizos28,29. As expected, Sonoran children alleles were closer to those of Mexican Mestizos than those of other Latin American Mestizos. Only one of the 4 alleles that were compared (DQB1*0302) had a lower frequency for Sonoran children than for Mexican Mestizos. Additionally, there was a higher frequency for DQA1*0501 (p<0.05) in Sonoran children than in Latin American Mestizos. The highest allele frequencies in newborns from Sonora were for DQA1*0501 (0.233) and DQA1*0301 (0.232).
Allele frequencies of the HLA-DQ2 and HLA-DQ8 haplotypes in Sonoran newborns compared with different Mestizo and European ethnic groups.
| Mexican Mestizo | Latin American Mestizo | Basque | Spanish | Slovenian | ||
|---|---|---|---|---|---|---|
| Alleles | Sonoran newbornsn=397Frequency | Frequency range | Frequency range | n=83Frequency | n=301Frequency | n=70Frequency |
| DQA1*0501 | 0.233a | 0.222-0.227a | 0.091-0.169b | 0.260a | 0.223a | 0.429c |
| DQB1*0201 | 0.132a | 0.163-0.167a | 0.125a | 0.480b | 0.462b | 0.329b |
| DQA1*0301 | 0.232a | 0.257-0.296a | 0.258-0.295a | 0.127b | 0.166b | 0.129b |
| DQB1*0302 | 0.105a | 0.237-0.278b | 0.242-0.295b | 0.067a | 0.193b | 0.143a |
In the last columns, Table 1 shows the comparison of Sonoran newborns with general Spanish and Basque populations; a Slovenian population was used as a Caucasian reference. There were no significant differences (p>0.05) among Sonoran newborns and Basques or Spaniards for DQA1*0501 allele frequency, as reported by Sánchez-Velasco et al.30 and Planelles et al.31 The low frequency of the DQB1*0302 allele for Sonoran newborns in this study was only comparable (p>0.05) to the allele frequency of Basques30 and Slovenians. There was absolutely no other similarity between Sonoran children and Slovenians15 in relation to any of the remaining 3 alleles analyzed.
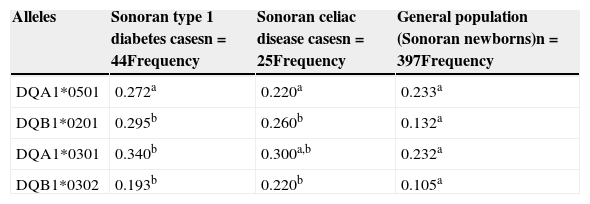
Regarding the T1D and CD Sonoran cases, we found a similar frequency in the alleles studied, when comparing the 2 groups (p>0.05). However, a comparison of these groups with data from Sonoran newborns, showed that T1D and CD patients had an allele frequency of DQB1*0201 and DQB1*302 that was significantly higher (p<0.05) than that of the general population, as shown in Table 2. Likewise, DQA1*0301 was the most prevalent risk allele in T1D and CD patients. Nevertheless, the higher frequency was only significant (p<0.05) when the T1D group was compared with the newborns from the general population, but not for the CD group.
Allele frequencies of the HLA-DQ2 and HLA-DQ8 haplotypes in Sonoran T1D and CD cases compared with the general population (Sonoran newborns).
| Alleles | Sonoran type 1 diabetes casesn=44Frequency | Sonoran celiac disease casesn=25Frequency | General population (Sonoran newborns)n=397Frequency |
|---|---|---|---|
| DQA1*0501 | 0.272a | 0.220a | 0.233a |
| DQB1*0201 | 0.295b | 0.260b | 0.132a |
| DQA1*0301 | 0.340b | 0.300a,b | 0.232a |
| DQB1*0302 | 0.193b | 0.220b | 0.105a |
n: sample size.
The different superscripts indicate significant differences (p<0.05) between the general population, T1D cases, and CD cases.
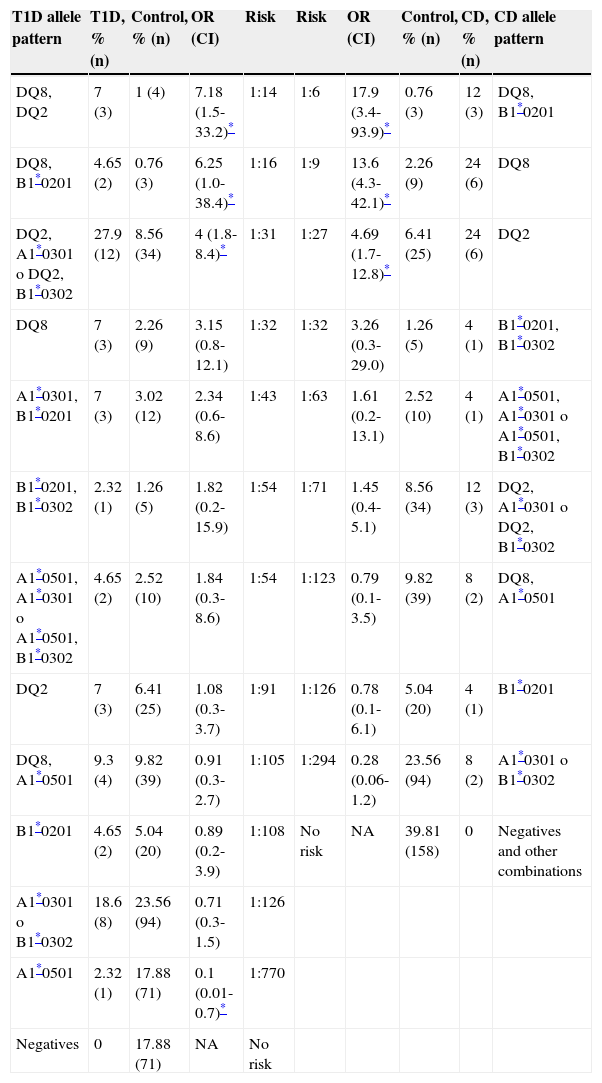
Based on the frequencies, a risk gradient was developed for T1D and CD in the Sonoran population, according to the different combinations of the 4 HLA-DQ2 and DQ8 alleles studied (Table 3). Our findings show that the CD group was more homogeneous, resulting in fewer positive risk allele combinations than those found in the T1D group. Therefore, the T1D patients presented with the 16 possible allele combinations, whereas we found only 13 in the CD group. The remaining 3 combinations were grouped as “other” for CD analysis purposes, to complete the 100% of the control group.
Disease risk gradient. T1D and CD risks were evaluated from HLA-DQ allele frequencies, considering a disease prevalence of 1:300 for T1D and 1:100 to 1:200 for CD, highlighted in the table.
| T1D allele pattern | T1D, % (n) | Control, % (n) | OR (CI) | Risk | Risk | OR (CI) | Control, % (n) | CD, % (n) | CD allele pattern |
|---|---|---|---|---|---|---|---|---|---|
| DQ8, DQ2 | 7 (3) | 1 (4) | 7.18 (1.5-33.2)* | 1:14 | 1:6 | 17.9 (3.4-93.9)* | 0.76 (3) | 12 (3) | DQ8, B1*0201 |
| DQ8, B1*0201 | 4.65 (2) | 0.76 (3) | 6.25 (1.0-38.4)* | 1:16 | 1:9 | 13.6 (4.3-42.1)* | 2.26 (9) | 24 (6) | DQ8 |
| DQ2, A1*0301 o DQ2, B1*0302 | 27.9 (12) | 8.56 (34) | 4 (1.8-8.4)* | 1:31 | 1:27 | 4.69 (1.7-12.8)* | 6.41 (25) | 24 (6) | DQ2 |
| DQ8 | 7 (3) | 2.26 (9) | 3.15 (0.8-12.1) | 1:32 | 1:32 | 3.26 (0.3-29.0) | 1.26 (5) | 4 (1) | B1*0201, B1*0302 |
| A1*0301, B1*0201 | 7 (3) | 3.02 (12) | 2.34 (0.6-8.6) | 1:43 | 1:63 | 1.61 (0.2-13.1) | 2.52 (10) | 4 (1) | A1*0501, A1*0301 o A1*0501, B1*0302 |
| B1*0201, B1*0302 | 2.32 (1) | 1.26 (5) | 1.82 (0.2-15.9) | 1:54 | 1:71 | 1.45 (0.4-5.1) | 8.56 (34) | 12 (3) | DQ2, A1*0301 o DQ2, B1*0302 |
| A1*0501, A1*0301 o A1*0501, B1*0302 | 4.65 (2) | 2.52 (10) | 1.84 (0.3-8.6) | 1:54 | 1:123 | 0.79 (0.1-3.5) | 9.82 (39) | 8 (2) | DQ8, A1*0501 |
| DQ2 | 7 (3) | 6.41 (25) | 1.08 (0.3-3.7) | 1:91 | 1:126 | 0.78 (0.1-6.1) | 5.04 (20) | 4 (1) | B1*0201 |
| DQ8, A1*0501 | 9.3 (4) | 9.82 (39) | 0.91 (0.3-2.7) | 1:105 | 1:294 | 0.28 (0.06-1.2) | 23.56 (94) | 8 (2) | A1*0301 o B1*0302 |
| B1*0201 | 4.65 (2) | 5.04 (20) | 0.89 (0.2-3.9) | 1:108 | No risk | NA | 39.81 (158) | 0 | Negatives and other combinations |
| A1*0301 o B1*0302 | 18.6 (8) | 23.56 (94) | 0.71 (0.3-1.5) | 1:126 | |||||
| A1*0501 | 2.32 (1) | 17.88 (71) | 0.1 (0.01-0.7)* | 1:770 | |||||
| Negatives | 0 | 17.88 (71) | NA | No risk |
CI: confidence interval; NA: not available; OR: odds ratio.
For T1D, we obtained a risk gradient of 1:14-1:126. Similarly, for CD, the calculated gradient ranged from 1:6-1:126. The 3 highest risks for T1D were encoded by HLA-DQ8 in combination with DQ2 or one of its alleles, or DQ8 alone. The 2 highest risks for CD were related to HLA-DQ8, as HLA-DQ8/DQB1*0201 or DQ8 alone, while the third one was HLA-DQ2. The OR and confidence intervals for these allele combinations are shown in Table 3.
According to the data obtained, the lower risk allele accounted for in this population was the DQA1*0501 (OR 0.1, p=0.02).
In relation to serologic screening, 42/44 patients in the diabetes case panel presented with positive anti-gliadin IgG antibodies and of these, 8 were positive for anti-gliadin IgA and 3 of them for anti-tTG IgA antibodies, as well. In the CD group, 16/25 cases were positive for anti-gliadin IgA and 15 had anti-tTG IgA at the time of analysis, indicating active CD.
DiscussionDistinctive Sonoran allele frequency is in all likelihood due to the blending of native population groups that were quite isolated from Mesoamerica and the first Europeans coming to the Northwest region of Mexico. The first immigrants were most probably people that worked in the mines in the eastern mountains of the current State of Sonora more than 300 years ago. Migrants were mainly Spaniards from Andalucía, Extremadura, and the Basque regions32, and they admixed with the local native groups of Mayos, Yaquis, Seris, Pimas, and the Tohono O’odham Nation33. Moreover, some additional blending may have occurred through the more recent immigration of people from central and southern Mexico, due to agricultural and industrial regional development. Thus, the State of Sonora has generated a new population with a complex genetic structure that has a higher European and lower Amerindian ancestry compared with other Mexican Mestizos20.
The major histocompatibility complex, or HLA in humans, is one of the most polymorphic regions in the human genome34. Therefore, haplotype frequency presents a wide variation in different populations worldwide. The DQ2 frequency in Caucasian populations from Western Europe has been estimated at 20-30%, while HLA-DQ8 frequency is common in South and Central America; approximately 20% of the Amerindians carry DQ835.
However, there is a worldwide similarity in the frequencies of these alleles in patients with T1D and CD. For example, the results presented for T1D match those of Swedish T1D patients (n=430)36; their frequencies for the DQA1*0501 (0.272) and DQB1*0201 (0.281) alleles are statistically equal to those of the Mexicans in this study. Regarding the DQ8-associated alleles, Scandinavian T1D patients36 carried significantly higher frequencies for the DQA1*0301 (0.403) and DQB1*0302 (0.364) alleles than the Sonoran TD1 children. Nevertheless, DQA1*0301 was the most prevalent allele in our T1D sample (0.340).
The distribution of alleles related to the HLA-DQ2 and DQ8 haplotypes has clinical relevance. Both DQB1*0201 and *0302 alleles had the greatest increase in prevalence in our T1D and CD sample, doubling the frequency in Sonoran newborns. This finding correlates with the risk gradient for both diseases, in which the highest risk combinations include the DQB1 fractions, as previously described for these diseases in other populations37.
For example, people of Iranian origin have the genotype DQB1*0201/x or DQB1*0302/x in 98% of confirmed CD cases. Moreover, in these Iranian cases, the DQB1*0201 allele was more prevalent in patients with severe atrophy and intestinal mucosal damage assessed by biopsy, presenting a Marsh IIIa-c grade38. In the present study, four of the CD cases had a Marsh III gut biopsy result39, and 3 of them were positive for the risk allele DQB1*0201.
Meanwhile, in T1D, the presence of DQB1 alleles has been associated with a younger age at diagnosis in a European population40 and the presence of multiple autoantibodies in U.S. Hispanics41. In our Sonoran sample, the presence of both alleles was associated with the development of T1D before 15 years of age (p<0.05).
Although there were no differences between the T1D and CD groups in the allele frequency analysis, they arose when we assessed the risk gradient. This is because even though the proportion was similar, the way that these alleles were distributed and combined varied from one disease to another. Thus, in this population the presence of the DQ2 haplotype alone accounted for a moderate T1D risk (1:91), whereas it represented a high risk for CD (1:27).
However, contrary to expectations, the HLA-DQ8 was more prevalent than the DQ2 in the Sonoran CD cases. This can be attributed to the special distribution of the risk alleles in the Sonoran population, in which DQ8 has a similar prevalence to that of DQ2. As a result, DQ8 is more frequent in Sonoran than in European populations, so the possibility of developing the disease increases, despite the fact that physiologically, DQ8 has a lower affinity than DQ2 for binding immunogenic gluten peptides12.
The highest risk combination for T1D was HLA-DQ2/DQ8, whereas we could not establish this association for the CD group, probably due to the sample size. However, this combination also confers a very high risk for CD development42. A striking finding was that the presence of this rare combination of haplotypes in the Sonoran general population increased the possibility of developing T1D to 10%, regardless of other associated factors. Additionally, according to our results, that particular situation was more common in the T1D cases than in the CD ones.
Concerning the T1D patients, the OR of the DQ2/DQ8 genotype (7.18) was higher than the sum of the OR of the DQ2 and DQ8 haplotypes (4.23). This supports the reported synergic effects between these haplotypes36.
Nevertheless, to have a better estimate of the actual risk, it would be necessary to consider situations in which the person is heterozygous or homozygous for a given haplotype, as well as for the HLA-DQ2.2 alleles that were not analyzed. The antigen-presenting cells from individuals who are homozygous for the HLA susceptibility alleles present gliadin-derived peptides more efficiently than heterozygotes, correlating with disease risk12. In children from The Environmental Determinants of Diabetes in the Young (TEDDY) study, a cumulative risk for CD of 11% was thus found among homozygous children for the DR3-DQ2 haplotype. This represents a greater risk than the 3% estimated risk among those with the DR3–DQ2/DR4–DQ8 haplotype43.
Our results may have interesting applications for identifying and estimating the risk for these diseases in Sonoran special groups, such as the relatives of the patients, or those with autoimmune disorders or associated environmental risk factors. In recent years, the European Society for Pediatric Gastroenterology, Hepatology, and Nutrition (ESPGHAN) working group has recommended performing HLA haplotype typing to add strength to the diagnosis of CD in symptomatic children. This approach is suggested for its high negative predictive value44 when small bowel biopsies cannot be performed and specific antibody titers are high.
While in more than 90% of the cases the development of CD requires the HLA DQ2 or DQ8 haplotypes, some T1D cases also carried other genetic variants35. Thus, as recommended by the American Diabetes Association, together with the measurement of CD-associated autoantibodies, haplotype analysis will be useful as a screening tool in patients with T1D, even in the absence of typical signs and symptoms1.
In this study, we evaluated anti-tTG and anti-gliadin antibody screening in children with diabetes, identifying 3 possible cases of CD in our T1D group, representing 7% of our diabetic sample. These results are below the 10.7% previously reported in Mexican mestizos with T1D45, perhaps because the T1D patients evaluated were incidental cases, and the risk for developing CD remains high throughout the lifetime of T1D patients. With the present approach, we found that some of the T1D cases had IgA anti-gliadins antibodies. This was probably due to the fact that these children went through an acute process in which intestinal permeability was compromised, creating antibodies against gliadins, and not necessarily because of CD. Hence this screening is a minimally invasive cost-effective option for periodically monitoring T1D patients and their risk for developing CD. This would opportunely detect possible CD cases, avoiding complications related to late diagnosis. In this context, and especially in asymptomatic patients, an endoscopy and biopsy assessment to confirm the CD diagnosis is recommended.
In summary, T1D and CD are diseases that frequently appear together. Since the Sonoran population has special genetic features due to its ancestry, the T1D and CD haplotype distribution was different from that reported in Europe, with a HLA-DQ2: HLA-DQ8 ratio of 1.2:1. The allele combination that conferred the greatest risk for T1D in the Sonoran population was the HLA-DQ8/DQ2 combination, followed by DQ8 alone or combined with DQB1*0201, whereas the DQ8/DQB1*0201 combination had the highest risk for CD.
Ethical responsibilitiesProtection of persons and animalsThe authors declare that the procedures followed conformed to the ethical standards of the responsible committee on human experimentation and were in accordance with the World Medical Association and the Declaration of Helsinki.
Data confidentialityThe authors declare that they have followed the protocols of their work center in relation to the publication of patient data.
Right to privacy and informed consentThe authors have obtained the informed consent of the patients and/or subjects referred to in the article. This document is in the possession of the corresponding author.
Financial disclosureFinancial support was received from the Consejo Nacional de Ciencia y Tecnología (CONACYT), grant S0008-2009-01-115212.
Conflict of interestThe authors declare that there is no conflict of interest.
The authors wish to thank Dr. N. Sotelo-Cruz, Dr. J.B. Elizondo, and Dr. G. García for their diagnoses and L. López-Domínguez, S. Aguayo-Patrón, and J.R. Valenzuela for their technical assistance.
Please cite this article as: Mejía-León ME, Ruiz-Dyck KM, Calderón de la Barca AM. Gradiente de riesgo genético HLA-DQ para diabetes tipo 1 y enfermedad celíaca en el noroeste de México. Revista de Gastroenterología de México. 2015;80:135–143.