Celiac disease (CD) is a complex condition, whose main genetic determinant involves HLA molecules, specifically the HLA-DQ2 and/or HLA-DQ8 heterodimers. Nevertheless, the frequency of the alleles encoding those molecules has not been reported in Venezuelan celiac patients. Therefore, the aim of our study was to evaluate the frequency of the HLA-DQB1 alleles in individuals with symptoms suggestive of CD and define the diagnostic markers of the condition in a Venezuelan population.
Material and methodsA cross-sectional study included 516 individuals with symptoms suggestive of CD. Molecular typing of the HLA-DQB1 locus was performed using a polymerase chain reaction-sequence-specific oligonucleotide procedure (PCR-SSO).
ResultsA total of 58.3% of the individuals with clinical manifestations consistent with CD presented with at least one risk allele (DQB1*0201 and/or DQB1*0302), and the diagnosis was confirmed in 40 of them. The patients with CD had a higher frequency of the DQB1*0201 risk allele (26.25%), followed by the DQB1*0302 (17.5%) allele. There was an association between the presence of risk alleles and the presence of lesions characteristic of CD (P = 0.001), and a correlation was found between the genetic predisposition to develop CD and the presence of anti-tissue transglutaminase antibodies (P = 0.0127).
ConclusionsThe results support the role of the DQB1*02 and DQB1*0302 alleles in CD susceptibility and the histologic alterations of the intestinal mucosa, in a Venezuelan population.
La enfermedad celíaca (EC) es una condición compleja cuyo principal determinante genético implica las moléculas HLA, específicamente los heterodímeros HLA-DQ2 y/o HLA-DQ8. No obstante, la frecuencia de los alelos codificantes de estas moléculas no ha sido reportada en pacientes celíacos venezolanos; por lo tanto, el objetivo de nuestro estudio fue evaluar la frecuencia de los alelos HLA-DQB1 en individuos con clínica sugestiva de EC y definir los marcadores de diagnóstico de esta condición en la población venezolana.
Material y métodosEstudio transversal, que incluyó 516 individuos con clínica sugestiva de EC. La tipificación molecular del locus HLA-DQB1 se realizó con la técnica de reacción en cadena de la polimerasa e hibridación con oligonucleótidos de secuencias específicas (PCR-SSO).
ResultadosEl 58.3% de los individuos con manifestaciones clínicas compatibles con EC presentaban por lo menos un alelo de riesgo (DQB1*0201 y/o DQB1*0302), y en 40 de ellos se confirmó el diagnóstico. Los pacientes con EC presentaron una mayor frecuencia del alelo de riesgo DQB1*0201 (26.25%), seguido por el alelo DQB1*0302 (17.5%). Se observó la asociación entre la presencia de alelos de riesgo y la presencia de lesiones características de la EC (p = 0.001) y se halló una correlación entre la predisposición genética a desarrollar EC y la presencia de anticuerpos anti-transglutaminasa (p = 0.0127).
ConclusionesLos resultados apoyan el papel de los alelos DQB1*02 y DQB1*0302 en la susceptibilidad y las alteraciones histológicas de la mucosa intestinal de la EC en la población venezolana.
Celiac disease (CD) is an autoimmune disorder, in which the key genetic factors (human leukocyte antigen [HLA]-DQ2 and HLA-DQ8), the autoantigen involved (tissue transglutaminase [tTG]), and the environmental trigger (gluten) have been well-defined.1 The condition is characterized clinically, by different intestinal and extraintestinal manifestations; histologically, by damage to the intestinal mucosa; and serologically, by the presence of anti-tissue transglutaminase 2 (anti-tTG2), anti-endomysial, and/or deamidated gliadin peptide antibodies.2
As is well-known, the main genetic determinant in CD involves HLA molecules, specifically the HLA-DQ2 and/or HLA-DQ8 heterodimers.3 The HLA-DQ2 heterodimer is composed of an α subunit and a β subunit, encoded by the HLA-DQA1*05 and HLA-DQB1*02 alleles, respectively, differentiating two types of DQ2 heterodimers: DQ2.5 (DQA1*0501/DQB1*0201) and DQ2.2 (DQA1*0201/DQB1*0202). The HLA-DQ8 heterodimer is composed of an α chain and a β chain, encoded by the HLA-DQA1*0301 and HLA-DQB1*0302 alleles, respectively.4 Those HLA-DQ molecules play a key role in CD due to their physicochemical properties and the binding of specific peptides that have been deamidated by the tissue transglutaminase 2 enzyme (tTG2), triggering an innate and adaptive immune response against gluten. Neutrophils, eosinophils, mastocytes, and complement proteins are activated in the innate immune response, potentially contributing to the pathogenesis of the disease. In the adaptive immune response, peptides derived from gluten and modified by tTG2 are presented by antigen-presenting cells in the mesenteric lymph nodes to CD4 + T cells, in the context of HLA-DQ2 and/or DQ8. The resulting Th1-type response leads to IFN-γ production and intestinal inflammation. Chronic inflammation conduces to the expansion and persistence of γδ T cells, which also contribute to IFN-γ production. In addition, gluten peptides induce IL-15 and stress molecule expression on enterocytes. The elevated IL-15 levels promote a NK-like phenotype in CD8 + T cells, directly contributing to enterocyte death. Furthermore, plasma cells are also abundant in the lesion, some of which express the immunodominant gluten peptide and are induced to secrete antibodies that bind to tTG2 and other targets. However, other less well-characterized mechanisms may participate in lesion development.5 Considering that CD is a complex disorder that involves both genetic and environmental factors, as well as the deregulation of the immune response, the aim of our study was to evaluate the frequency of the HLA-DQB1 alleles in individuals with symptoms suggestive of CD and define the diagnostic markers of the condition in a Venezuelan population.
Materials and methodsPatientsAn exploratory and correlational study was conducted that included 516 Venezuelan mestizo individuals from different health centers in the Capital District of Venezuela. They went to the Immunogenetic Section of the Experimental Medicine Center of the Instituto Venezolano de Investigaciones Científicas (IVIC) to undergo the molecular determination of HLA-DQB1 alleles because they had symptoms suggestive of CD.
The Venezuelan mestizo is defined as someone born in Venezuela that is a mixture of Amerindian and European (mainly Spanish) ancestry.
The clinical and serologic characteristics and duodenal biopsy results were registered at the time of HLA-DQB1 genotyping. Included in the study were individuals with general manifestations (fatigue, weight loss, slowed growth), gastrointestinal manifestations (dyspepsia, nausea, vomiting, abdominal pain, diarrhea, constipation), and the presence of extraintestinal manifestations (cutaneous manifestations).
CD diagnosis was based on the “four-out-of-five rule”, through which four of the five following criteria are sufficient for diagnosing the condition: (1) typical signs and symptoms (diarrhea and malabsorption); (2) positive antibodies; (3) positivity for HLA-DQ2 and/or HLA-DQ8; (4) intestinal damage (villous atrophy and lesser lesions); and (5) clinical response to a gluten-free diet (GFD).6
Genetic studiesGenomic DNA was extracted from white blood cells, from venous blood draws, utilizing the Bunce protocol,7 and the polymorphism of the HLA-DQB1 locus was determined through the polymerase chain reaction (PCR) method and hybridization with specific-sequence oligonucleotide (SSO) probes, utilizing the Dynal RELI™ SSO HLA- DQB1 kit (Dynal Biotech Ltd, Invitrogen, United Kingdom).
Histologic studiesThe duodenal biopsy samples were histopathologically analyzed by an anatomopathologist and the mucosal damage was evaluated, utilizing the Marsh classification, as: type 0, normal mucosa; type 1, infiltrative lesion; type 2, hyperplastic lesion; and type 3, destructive lesion, subcategorized as type 3a-mild (partial villous atrophy), type 3b-marked (subtotal villous atrophy), and 3c-total (total villous atrophy).
Classification of the individuals with symptoms suggestive of CDBased on the HLA-DQB1 genotypes, i.e., in the absence or presence of one or two risk alleles (HLA-DQB1 *0201, *0202, *0301, and *0302), in homozygous or heterozygous form, the individuals were classified into groups of no risk, low risk, intermediate risk, or high risk for presenting with CD.8
TreatmentAll the participating individuals were asked if they followed a GFD, which is the only treatment recommended for patients with CD. Said treatment was prescribed by the treating physician and monitored by a medical specialist in nutrition who evaluated treatment adherence (through a questionnaire that indicated the foods consumed by the patient) and CD antibody determination. The individuals that followed a GFD and who, after 6 months, showed clinical improvement and negative serology for anti-tissue transglutaminase antibodies, were considered diet responders.
Statistical analysisAllele and genotype frequencies were calculated through direct count. The differences in frequencies between groups were determined using the chi-square test and contingency tables, and statistical significance was set at a p value below 0.05. The odds ratio (OR), with the corresponding confidence interval (CI), was employed to calculate the associations between the risk alleles and disease occurrence and the presence of histologic lesions characteristic of CD. The correlations between the variability of the genes analyzed and the different variables were carried out using SPSS version 20 software,9 determining the Pearson’s correlation coefficient.
Ethical considerationsThe study protocol was approved by the institutional ethics committee (Record No. 4300) and carried out according to the guidelines of the Declaration of Helsinki. All participants signed written statements of informed consent.
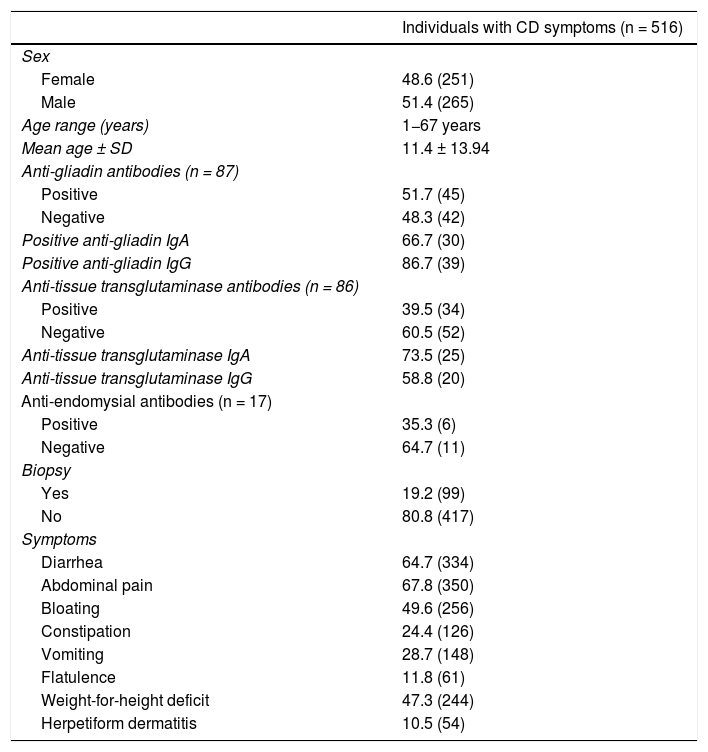
ResultsDemographic, clinical, and diagnostic characteristics of the individuals with symptoms suggestive of CDAll the individuals included in the study had symptoms related to CD and they included gastrointestinal manifestations (most frequently: diarrhea, constipation, dyspepsia, and flatulence) and non-gastrointestinal symptoms (most frequently: weight-for-height deficit and dermatitis, mainly in children). Table 1 shows the demographic and diagnostic characteristics and the main symptoms of the individuals with symptoms suggestive of CD. Of the 516 study individuals, 251 (48.6%) were females, 265 (51.4%) were males, 328 (63.6%) were adults, and 188 (36.4%) were under 18 years of age (age range: 1−67 years). Of the individuals with symptoms suggestive of CD, 87 (16.8%) had serologic testing (tissue transglutaminase IgA and IgG and anti-gliadin IgA and IgG) and 99 (19.2%) underwent duodenal biopsy.
Demographic, diagnostic, and symptomatologic characteristics of the individuals with symptoms suggestive of celiac disease.
| Individuals with CD symptoms (n = 516) | |
|---|---|
| Sex | |
| Female | 48.6 (251) |
| Male | 51.4 (265) |
| Age range (years) | 1−67 years |
| Mean age ± SD | 11.4 ± 13.94 |
| Anti-gliadin antibodies (n = 87) | |
| Positive | 51.7 (45) |
| Negative | 48.3 (42) |
| Positive anti-gliadin IgA | 66.7 (30) |
| Positive anti-gliadin IgG | 86.7 (39) |
| Anti-tissue transglutaminase antibodies (n = 86) | |
| Positive | 39.5 (34) |
| Negative | 60.5 (52) |
| Anti-tissue transglutaminase IgA | 73.5 (25) |
| Anti-tissue transglutaminase IgG | 58.8 (20) |
| Anti-endomysial antibodies (n = 17) | |
| Positive | 35.3 (6) |
| Negative | 64.7 (11) |
| Biopsy | |
| Yes | 19.2 (99) |
| No | 80.8 (417) |
| Symptoms | |
| Diarrhea | 64.7 (334) |
| Abdominal pain | 67.8 (350) |
| Bloating | 49.6 (256) |
| Constipation | 24.4 (126) |
| Vomiting | 28.7 (148) |
| Flatulence | 11.8 (61) |
| Weight-for-height deficit | 47.3 (244) |
| Herpetiform dermatitis | 10.5 (54) |
CD: celiac disease; Ig: immunoglobulin; SD: standard deviation.
Frequencies are shown in percentages and the number of individuals is in parentheses.
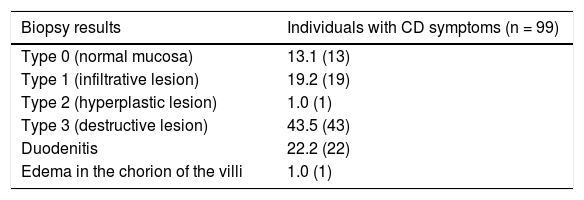
Table 2 shows the results of the biopsy taken in 99 individuals with symptoms suggestive of CD, 63 (63.6%) of whom presented with histologic findings consistent with CD. Utilizing the Marsh classification for mucosal damage, 13 (13.1%) individuals had normal mucosa (type 0), 19 (19.2%) had infiltrative lesion (type 1), one (1%) had hyperplastic lesion (type 2), and 43 (43.5%) had destructive lesion (type 3). Based on the grade of villous atrophy, 16 (37.2%) individuals had partial villous atrophy (3a), 8 (18.6%) subtotal villous atrophy (3b), and 19 (44.2%) total villous atrophy (3c).
Histologic findings in the intestinal biopsy performed on individuals with symptoms suggestive of celiac disease.
| Biopsy results | Individuals with CD symptoms (n = 99) |
|---|---|
| Type 0 (normal mucosa) | 13.1 (13) |
| Type 1 (infiltrative lesion) | 19.2 (19) |
| Type 2 (hyperplastic lesion) | 1.0 (1) |
| Type 3 (destructive lesion) | 43.5 (43) |
| Duodenitis | 22.2 (22) |
| Edema in the chorion of the villi | 1.0 (1) |
CD: celiac disease.
Frequencies are shown in percentages and the number of individuals is in parentheses.
Of the 516 individuals with symptoms suggestive of CD, only 77 (14.9%) followed a GFD, 35 (45.6%) of whom had a satisfactory clinical response, after following the diet. On the other hand, 43 (43.4%) of the individuals that underwent duodenal biopsy began a GFD, 28 (65.1%) of whom had gastrointestinal symptom improvement and negative serology, at month 6 of the diet.
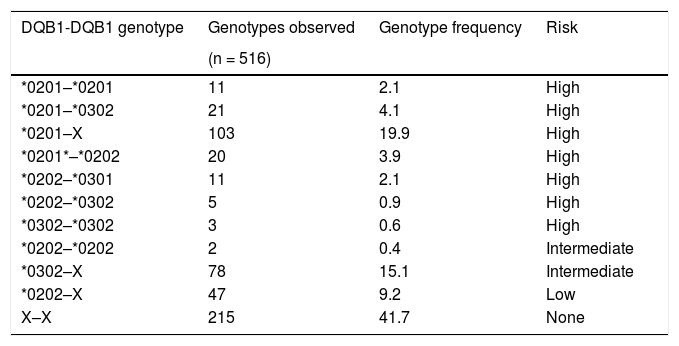
HLA-DQB1 polymorphismThe analysis of the distribution of the HLA-DQB1 genotypes showed that 301 (58.3%) of the individuals with symptoms suggestive of CD had risk genotypes. A total of 155 (51.5%) presented with at least one DQB1*0201 allele and 107 (35.5%) presented with at least one DQB1*0302 allele. Finally, 215 (41.7%) of the individuals with symptoms suggestive of CD did not present with a risk genotype (Table 3).
Distribution of the frequency of the HLA-DQB1 genotypes in individuals with symptoms suggestive of celiac disease.
| DQB1-DQB1 genotype | Genotypes observed | Genotype frequency | Risk |
|---|---|---|---|
| (n = 516) | |||
| *0201–*0201 | 11 | 2.1 | High |
| *0201–*0302 | 21 | 4.1 | High |
| *0201–X | 103 | 19.9 | High |
| *0201*–*0202 | 20 | 3.9 | High |
| *0202–*0301 | 11 | 2.1 | High |
| *0202–*0302 | 5 | 0.9 | High |
| *0302–*0302 | 3 | 0.6 | High |
| *0202–*0202 | 2 | 0.4 | Intermediate |
| *0302–X | 78 | 15.1 | Intermediate |
| *0202–X | 47 | 9.2 | Low |
| X–X | 215 | 41.7 | None |
X: HLA-DQB1 alleles different from the alleles associated with CD (HLA-DQB1 *0201, *0202, *0301, *0302).
Frequencies are shown in percentages.
In the study population, even though the frequency of the genotypes related to an increased risk for CD was 58.3% (301 individuals), the analysis of the 99 patients that underwent intestinal biopsy showed that only 54 (54.5%) had a risk genotype, confirming CD diagnosis in 40 (74.1%) of them. In the patients with CD, there was a higher frequency of the DQB1*0201 risk allele (0.2625), followed by the DQB1*0302 allele (0.175).
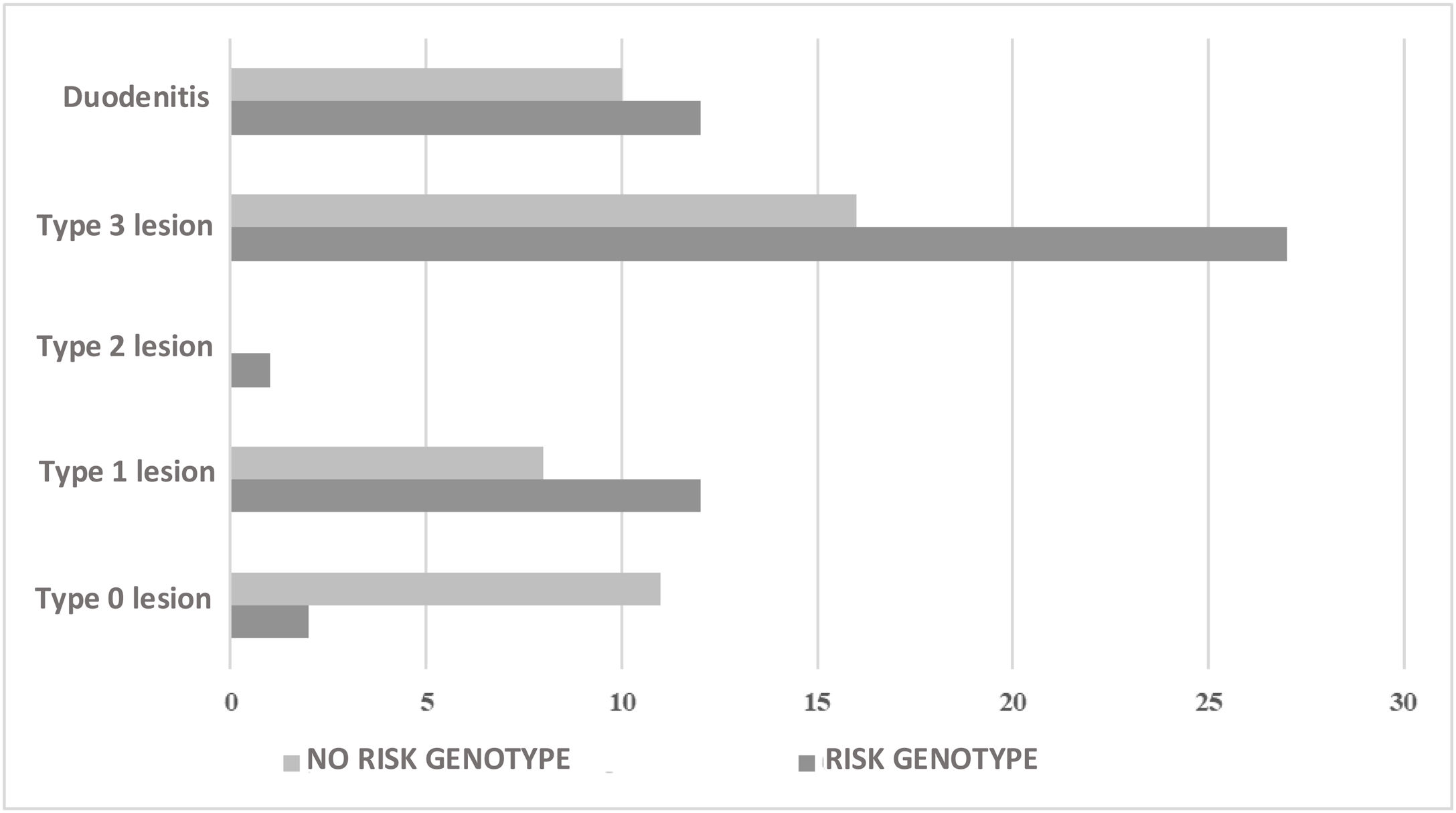
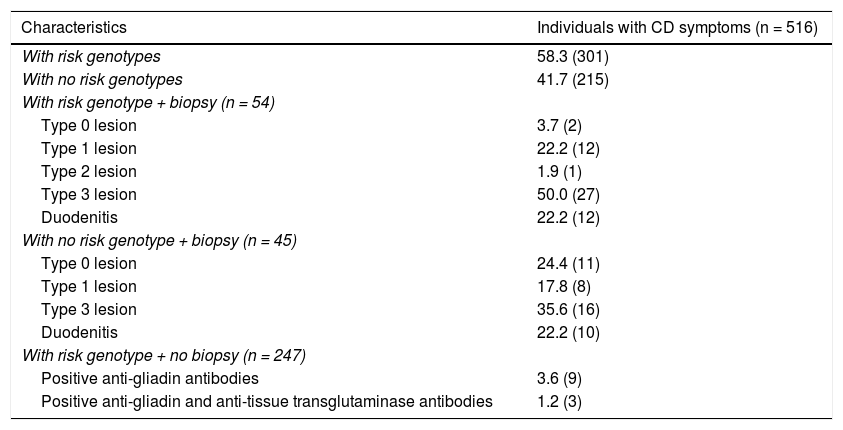
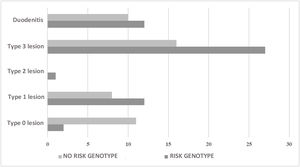
Of the 45 (45.5%) individuals that had no risk genotype, duodenal biopsy showed lesions characteristic of CD in 24 (53.3%) of them. With respect to the 417 individuals with no intestinal biopsy, 247 (59.2%) carried a risk genotype and 3 individuals had anti-gliadin IgA antibodies and anti-tissue transglutaminase IgA antibodies (Table 4). In the comparison of the individuals with biopsy, divided into groups with and without risk alleles, there was an association between the absence of risk alleles and the absence of mucosal lesions (OR: 0.11; 95% CI: 0.02−0.56; p = 0.001). Likewise, there was an association between the presence of risk alleles and the presence of lesions characteristic of CD, regardless of lesion type (OR: 9.2; 95% CI: 1.87–44.92; p = 0.001) (Fig. 1).
Histologic and serologic characteristics of the individuals with symptoms suggestive of celiac disease, classified according to the HLA-DQB1 genotype.
| Characteristics | Individuals with CD symptoms (n = 516) |
|---|---|
| With risk genotypes | 58.3 (301) |
| With no risk genotypes | 41.7 (215) |
| With risk genotype + biopsy (n = 54) | |
| Type 0 lesion | 3.7 (2) |
| Type 1 lesion | 22.2 (12) |
| Type 2 lesion | 1.9 (1) |
| Type 3 lesion | 50.0 (27) |
| Duodenitis | 22.2 (12) |
| With no risk genotype + biopsy (n = 45) | |
| Type 0 lesion | 24.4 (11) |
| Type 1 lesion | 17.8 (8) |
| Type 3 lesion | 35.6 (16) |
| Duodenitis | 22.2 (10) |
| With risk genotype + no biopsy (n = 247) | |
| Positive anti-gliadin antibodies | 3.6 (9) |
| Positive anti-gliadin and anti-tissue transglutaminase antibodies | 1.2 (3) |
CD: celiac disease.
Frequencies are shown in percentages and the number of individuals is in parentheses.
Upon establishing the correlations, one was found between the genetic predisposition to developing CD and the presence of anti-tissue transglutaminase IgG antibodies (r: 0.2574; p = 0.0127). Based on that correlation, the frequency of patients with CD and risk genotypes was compared between seropositive and seronegative individuals for anti-tissue transglutaminase antibodies (IgA and IgG), showing a significantly increased frequency in the seropositive patients with CD, compared with the seronegative patients (86.7% versus 57.9%, respectively; OR: 4.7; 95% CI: 0.825–27.069; p = 0.03).
DiscussionThe present study is the first to evaluate the frequency of HLA-DQB1 alleles in Venezuelan individuals with symptoms suggestive of CD.
The main findings included a positive association between the risk alleles (HLA-DQB1 *0201, *0302) and the presence of lesions in the intestinal mucosa, regardless of lesion type (infiltrative, hyperplastic, or destructive). Some studies have suggested that the HLA-DQB1*02 allele that encodes the DQ2 molecule determines the severity of damage to the intestinal mucosa, as well as its progression to possible complications. Specifically, patients with DQB1*02 homozygosity, not only present with more severe clinical symptoms, in particular, diarrhea and anemia, but also develop more severe intestinal lesions that take longer to heal, in the absence of a GFD.10 In our study, we found that 74% of the patients with villous atrophy had at least one DQB1*02 allele and 26% a DQB1*0302 allele, suggesting that the presence of the DQB1*02 and DQB1*0302 alleles, is associated not only with a predisposition to develop CD, but also with the presence of lesions in the intestinal mucosa. Nevertheless, the patients diagnosed with CD that followed a GFD showed clinical improvement and none of them presented with intestinal lymphoma or refractory CD.
Utilizing the criteria of the “four-out-of-five rule”, which includes clinical manifestations, serology, risk genotype, intestinal biopsy results, and clinical response to a GFD, we confirmed the diagnosis of CD in 40 of the individuals with symptoms suggestive of CD. By comparing the frequencies of the HLA-DQB1 alleles in the patients with CD diagnosis and the frequencies described in celiac patients from other populations, we found an increased frequency of the DQB1*0201 and DQB1*0302 risk alleles, similar to that described in a European population11 and in Latin American mestizo populations, such as those in Argentina,12,13 Brazil,14–17 Chile,18,19 Uruguay,20 and Cuba,21 populations characterized by having a White component above 50%. On the other hand, we found a lower frequency of the HLA-DQB1*0302 allele, with respect to the frequency described in populations with a higher prevalence of Amerindian genes.22,23 Recently, Mejía-León and Calderón-de la Barca24 highlighted the fact that, in the Mexican population, the DQ2 and DQ8 molecules contributed to the genetic risk for CD, showing that the risk was not exclusively due to DQ2, as in Europeans, or to DQ8, as seen in populations with a greater prevalence of Amerindian genes.
In individuals with no risk genotypes and with symptoms suggestive of CD, the presence of intestinal lesions characteristic of CD could be explained by the existence of other causes of intestinal villous atrophy, including autoimmune enteropathy, variable common immunodeficiency, enteropathy associated with olmesartan, lymphoproliferative disorders of the small bowel, and atrophy of infectious origin,25 as well as the presence of other non-HLA variants associated with CD. Association studies on the complete genome have identified more than 40 non-HLA genes associated with CD,26,27 that are relevant, even if they confer a limited genetic risk, because they enable the discovery of potential key pathways involved in the pathogenesis of the disease.
One percent of the individuals with symptoms suggestive of CD, that carried a risk genotype, but did not undergo intestinal biopsy, presented with anti-gliadin IgA antibodies and anti-tissue transglutaminase IgA antibodies, as well as with clinical improvement upon following a GFD. That group was made up only of children, with a mean age of 3.6 ± 2.8 years, and could be classified as patients with a presumptive diagnosis of CD, despite not having undergone intestinal biopsy. Even though there is an ongoing debate for defining whether or not noninvasive tests can be used as the only tests for diagnosing CD, positivity for two different serologic tests and the presence of risk genotypes (HLA-DQ2 and/or HLA-DQ8) in children has been suggested as a possible test in the diagnostic algorithm for CD, thus avoiding the performance of gastrointestinal endoscopy with biopsy.3
Finally, individuals with risk genotypes and the presence of anti-tissue transglutaminase antibodies (IgA-IgG) are 4.7-times more prone to develop CD, confirmed upon observing an increased frequency of individuals diagnosed with CD, in the group of patients with risk genotype and seropositivity for anti-tissue transglutaminase antibodies. Even though CD-specific antibodies are utilized for the presumptive diagnosis of CD, it has been suggested that those antibodies carry out important functions in relation to disease progression, at the level of the small bowel mucosa, and in the development of extraintestinal manifestations related to CD.
ConclusionsThere is scant research on CD in the Venezuelan population, and in that sense, one of the main contributions of the present study is that the results support the role of the DQB1*02 and DQB1*0302 alleles in disease susceptibility and in the histologic alterations of CD in that population. Therefore, the genotyping of those specific genes in the Venezuelan population would be useful for screening subjects at risk for CD, distinguishing individuals with a low probability for developing the disease from individuals that should be monitored for possible future development of the disorder.
Financial disclosureNo financial support was received in relation to this study/article.
Conflict of interestThe authors declare that there is no conflict of interest.
Please cite this article as: Fernández-Mestre M, Padrón-Lowe D, Salazar-Alcalá E, Blanco-Pérez F. Papel de los alelos HLA-DQB1 en el riesgo, signos y síntomas y gravedad de la enfermedad celíaca en la población venezolana. Revista de Gastroenterología de México. 2023;88:125–131.