Gut microbiota is the community of live microorganisms residing in the digestive tract. There are many groups of researchers worldwide that are working at deciphering the collective genome of the human microbiota. Modern techniques for studying the microbiota have made us aware of an important number of nonculturable bacteria and of the relation between the microorganisms that live inside us and our homeostasis. The microbiota is essential for correct body growth, the development of immunity, and nutrition. Certain epidemics affecting humanity such as asthma and obesity may possibly be explained, at least partially, by alterations in the microbiota. Dysbiosis has been associated with a series of gastrointestinal disorders that include non-alcoholic fatty liver disease, celiac disease, and irritable bowel syndrome. The present article deals with the nomenclature, modern study techniques, and functions of gut microbiota, and its relation to health and disease.
La microbiota intestinal es la comunidad de microorganismos vivos residentes en el tubo digestivo. Muchos grupos de investigadores a nivel mundial trabajan descifrando el genoma de la microbiota. Las técnicas modernas de estudio de la microbiota nos han acercado al conocimiento de un número importante de bacterias que no son cultivables, y de la relación entre los microorganismos que nos habitan y nuestra homeostasis. La microbiota es indispensable para el correcto crecimiento corporal, el desarrollo de la inmunidad y la nutrición. Las alteraciones en la microbiota podrían explicar, por lo menos en parte, algunas epidemias de la humanidad como el asma y la obesidad. La disbiosis se ha asociado a una serie de trastornos gastrointestinales que incluyen el hígado graso no alcohólico, la enfermedad celíaca y el síndrome de intestino irritable. En el presente trabajo trataremos sobre la nomenclatura, las técnicas de estudio modernas, las funciones de la microbiota intestinal y la relación que tiene con la salud y la enfermedad.
Our knowledge of the interesting relationship between human beings and the microorganisms we harbor has greatly increased over the past years. We no longer call these living entities «intestinal flora», nor do we regard them as simply commensal. In fact, we humans are «super organisms» governed in part by the microorganisms living inside us.1 The aim of this review is to familiarize the reader with the current terms used in the thriving field of the human microbiota, in particular the gut microbiota, to know the profound implications of diet and the environment on the normal and abnormal microbiota, and to outline a panorama of the relation between the microbiota and gastrointestinal diseases.
The literature review was carried out by consulting the PubMed database of information encompassing the last 15 years, as well as the studies presented at the 2012 Digestive Diseases Week in San Diego, California, and the 2012 United European Gastroenterology Week in Amsterdam.
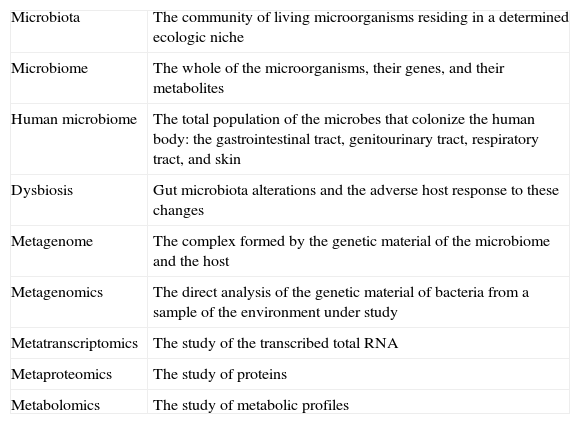
Microbiota and other conceptsIt is worthwhile to become familiar with a series of terms that are currently employed in this field. The term microbiota refers to the community of living organisms residing in a determined ecologic niche. The microbiota living in the human gut is one of the most densely populated communities,2 surpassing that of the soil, the subsoil, and the oceans. In the mammalian large intestine the number of microorganisms reaches 1012-1014, even more than the number of human cells.3 The microbial ecosystem of the intestine (gut microbiota) includes many native species that permanently colonize in the gastrointestinal tract and a variable series of microorganisms that only do so transitorily. The whole of the microorganisms, their genes, and their metabolites is called the microbiome. The human microbiome refers to the total population of microbes colonizing the human body, including the gastrointestinal tract, genitourinary tract, oral cavity, nasopharynx, respiratory tract, and skin.4 The Human Microbiome Project has identified approximately 30% of the gut microbiota5 and together with the Metagenomics of the Human Intestinal Tract in Europe and many other groups, is actively working to identify all of the genes of the microbiota.
Dysbiosis is defined as the alterations in the gut microbiota and the adverse response of the host to these changes. It has been associated with diseases as dissimilar as asthma, chronic inflammatory disease, obesity, and non-alcoholic steatohepatitis (NASH). 6–8
There have been several challenges involved in the study of the microbiome in the past: not all the microorganisms are easy to grow. Nevertheless, the modern techniques for studying genetic material have revolutionized our understanding of the microbiome. Some components of the microbiota require special conditions for their growth in culture media and therefore they went undetected or were unknown in the past. For example, the colonic microbiota have approximately 800 to 1,000 species per individual, but 62% of them were unknown and 80% of the bacteria identified by metagenomics are regarded as unculturable.9
The concepts and advances in «metanomics» have opened a window into the understanding of the gut microbiota 10 (Table 1):
- •
Metagenomics is the analysis of the genetic material of bacteria taken directly from a sample of the environment that is being studied, making it possible to identify bacteria that cannot be detected in culture media.
- •
Metatranscriptomics studies the transcribed total RNA.
- •
Metaproteomics focuses on protein levels.
- •
Metabolomics studies metabolic profiles.
- •
The metagenome is the complex formed by the host and the microbiome.
Concepts of microbiota.
| Microbiota | The community of living microorganisms residing in a determined ecologic niche |
| Microbiome | The whole of the microorganisms, their genes, and their metabolites |
| Human microbiome | The total population of the microbes that colonize the human body: the gastrointestinal tract, genitourinary tract, respiratory tract, and skin |
| Dysbiosis | Gut microbiota alterations and the adverse host response to these changes |
| Metagenome | The complex formed by the genetic material of the microbiome and the host |
| Metagenomics | The direct analysis of the genetic material of bacteria from a sample of the environment under study |
| Metatranscriptomics | The study of the transcribed total RNA |
| Metaproteomics | The study of proteins |
| Metabolomics | The study of metabolic profiles |
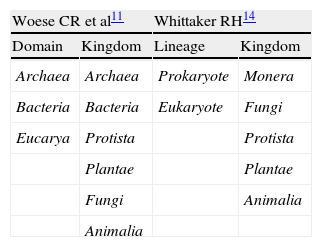
Various classification systems of the biologic kingdoms have been described (Table 2). In 1990 Woese introduced the term «domain» to substitute «kingdom» as the highest taxonomic order, dividing all living beings into Bacteria, Archaea, and Eucarya.11 The archaea, unicellular organisms formerly grouped in the bacteria domain, possess a sufficiently distinct genetic material from the bacteria to be classified in a separate domain.12 A recent discovery is the presence of members of the Archaea domain in the gut microbiota, currently regarded as distinct from the Bacteria domain. An example of the archaea is the methane-producing Methanobrevibacter smithii, and in recent studies it has been implicated in irritable bowel syndrome (IBS) with constipation.13
Ribosomal RNA (rRNA) is the most widely used macromolecule in bacterial phylogenetic and taxonomic studies.15 The sequencing of the variable regions of the gene that encodes for the 16S subunit of rRNA (16S rRNA) identifies the phylogenetic likeness of the bacteria and the archaea and enables them to be classified without the use of culture media. The genetic information obtained from the microbiome through the 16S rRNA is grouped into the so-called operational taxonomic units, according to the similarity percentage of their 16S rRNA. When there is a 95% similarity in the 16S rRNA, genus is being referred to, and when the similarity is 97%, the reference is to species.16
About 50% of the fecal mass is made up of bacteria. This population is composed of trillions of microorganisms that belong to 4 main phyla: Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria, with a predominance of the first two (90%).17
Functions of the microbiotaThe gut microbiota has gone from being considered an accompanying commensal to a «metabolic organ»,18 with functions in nutrition, immunity regulation, and systemic inflammation.19 Mammals that are raised germ free (GF) have an abnormal body development with intestinal wall atrophy, a low-weight heart, lungs and liver, and an immature immune system with low immunoglobulin levels.20 Backhed et al. showed that a group of mice had 40% more body fat than their GF counterparts while fed the same diet,21 and the GF mice were protected from obesity caused by high-fat and high-sugar diets.22 When microbiota was transplanted from the cecum of normal mice into the GF mice («conventionalizing»), there was a significant increase in their body fat content.21 The gut microbiota has enzymes that transform the complex polysaccharides of the diet that the human intestine cannot digest or absorb, into monosaccharides and short-chain fatty acids (SCFA), principally acetic, propionic, and butyric acid. The first two are absorbed into the portal circulation and the third is used by the colonocytes as a source of energy. The SCFA can be transported to the liver to be used in the synthesis of lipids; as a matter of fact, it is estimated that the calories derived from this bacterial digestion make up about 10% of all the energy we absorb.23 The quantity of SCFA in the colon and blood is important for host immunoregulation. Some studies report positive SCFA effects in patients with inflammatory alterations of the bowel; in fact, those patients have much lower SCFA concentrations.24–26 In addition, it appears that the microbiota is capable of modulating the genes that affect the disposition of energy in the adipocytes.2 The microbes and vertebrates evolved together over thousands of years and the normal functioning of the digestive and immunologic systems depends on the presence of the symbiotic microbiota.27
Factors that influence the microbiotaFrom an evolutionary perspective, the organisms that make up the microbiota in mammals are determined by the types of nutritional sources, and so omnivores, carnivores, and herbivores have different profiles.28 The characteristics of diet, together with genetic factors, influence the predominance of some microorganisms over others.29 After only one day of a Western diet (high in fat and sugar and low in plant polysaccharides), the mice showed changes in their microbial composition and their metabolic pathways and in 2 weeks they had developed greater adiposity.30 The abundance or scarcity of food will determine the presence or not of bacterial species that reproduce well when there is unlimited availability of food, or of the most efficient species when the nutrients are scare.29,31 Mice fed a Western diet show increased Firmicutes and reduced Bacteroidetes.30
In utero, the human being does not have a microbiota. Upon birth, the gastrointestinal tract colonizes immediately. Even the type of birth (natural or cesarean) and the type of food (breast milk or formula) have been shown to produce differences in the gut microbiota.32 Microbial fecal profiles of nursing infants show a marked similarity to the bacterial profiles of the birth canal and breast milk.33 During infancy and throughout life, the microbial composition also changes according to age and diet.34 In the first 2 years of life, the microbiota is dominated by biphidobacteria.35 Afterwards, the microbial composition diversifies and reaches its maximum complexity in the adult, with hundreds of phylotypes dominated by Bacteroidetes and Firmicutes.36
Even when the gut microbiota changes over the years, the environment and the maternal microbiota during birth and breast-feeding appear to remain very important factors in the future development of the microbiota. Once the microbiota is established in an individual, it is stable in relation to time.37 In humans, the bacterial communities are more similar among members of the same family than with unrelated individuals.38
Arumugam et al. recently postulated the concept of the enterotypes, with the idea of classifying the different microbiota of the human gut, based on the composition of their bacterial communities and according to the abundance of the diverse bacterial genuses.39 This could facilitate the association of the different enterotypes with the diverse conditions associated with dysbiosis. Studies such as these help to untangle the data that is available to us and to correlate the different microbial populations with the clinical entities that are the product of dysbiosis.
However, we must be careful when drawing conclusions and always think of the changes in the microbiota as the cause and not the consequence of the physiologic modifications of the organisms. For example, Purna et al. showed that the effects on the microbiota associated with the introduction of fiber into the diet could be related to bowel transit velocity and not to the fiber itself, given that the same changes were reproduced in the microbiota when using inert laxatives.40
Microbiota and immunityThe gut microbiota has an important effect on the human immune response. In 1989, Strachan suggested that the decrease in the microbial load due to improved standards of hygiene in the developed countries could lead to an increase in autoimmune diseases.41 Diet and its effects on the gut microbiota and on the immune response have been postulated as possible explanations for the increase in incidence of inflammatory diseases such as asthma and type I diabetes in the developed countries.27 New findings about the gut microbiota and its immunomodulatory capacity coincide with the epidemiologic data that connect obesity and asthma, or obesity and type I diabetes.42,43
The intestinal mucosa performs functions of adaptive immunity because its immune system has the capacity to respond to an infinite number of antigens, but there is also the innate immunity that is the recognition of specific antigens and is inherited phylogenetically from the plants to the vertebrates. These antigens have been called pathogen-associated molecular patterns (PAMPs) and they include lipids, lipopolysaccharides (LPS), and lipoproteins. The PAMPs are recognized by the pattern-recognition receptors (PRRs). The interaction between the PRRs and the PAMPs induces cytokine and interferon production. Among others, the PRRs include the Toll-like receptors (TLRs) that are transmembrane receptors. Various PAMPs that are ligands of the TLRs contain lipids, which are indispensible for their agonist activity, such as the bacterial LPS (bacterial endotoxins) that are TLR-4 ligands. The LPS are essential components of the bacterial cell wall. Even though they are not strictly factors of bacterial virulence, they awaken an intense innate immunologic response. The TLRs are expressed in the cells that are in charge of innate immunity, such as the macrophages, epithelial cells, endothelial cells, adipocytes, and in the parenchyma of some organs,44 but they are also expressed in the cells of the adaptive immunity cells that include B cells, mast cells, T cells, and the dendritic cells, which are key to the initiation of this adaptive immunity.44 The dendritic cells are a type of antigen-presenting cell. They are located in the lamina propria, they extend their appendages among the mucosal epithelial cells and display molecular patterns of pathogenic and commensal microorganisms.45 The signals arising from the TLRs induce the dendritic cells to differentiate and produce cytokines.46 The dendritic cells present the antigens to the T cells and are implicated in defense functions, as well as in immunologic tolerance to foods and microorganisms.47
When the LPS bind to the TLR-4, an intense inflammatory response is produced that damages the white tissues. The LPS are detected in the circulation of healthy individuals and their levels increase after the ingestion of energy-rich foods.48
Until a short while ago, adipose tissue was regarded as a mere storage compartment, but the adipocyte is an active adipokine-producing endocrine cell.49 In obesity, in addition to the increased adipocytic volume, the adipose tissue is infiltrated by macrophages. The macrophages have 2 subpopulations: M1 that produce inflammatory cytokines and M2 that generate anti-inflammatory products.50 The TLRs promote the M1 phenotype with the consequent increase in proinflammatory cytokines.
The «hygiene theory» supposes that the excess of cleanliness and the reduced exposure to bacteria at an early age impedes the correct development of the immunoregulatory mechanisms that prevent inappropriate T cell responses and the later inflammatory diseases.27 Hansen et al. demonstrated that GF mice that were «conventionalized» at the age of 3 weeks with the cecal content of normal mice, had permanently modified the composition of their gut microbiota and developed a proinflammatory immune response. In other words, the short postnatal germ-free period had permanent adverse effects on immunity.51 Interestingly, if the conventionalization is carried out at the first week of life, these effects are not reproduced, leading to the idea that there is a window of time in which immunity can be permanently modified.
There are radical differences between the gut microbiota of children in Africa and children in urban Europe. The children from Burkina Faso (Africa) have a diet that is very high in fiber and their microbiota has large quantities of Bacteroidetes that hydrolyze the complex plant polysaccharides, and they have a much lower abundance of Firmicutes than the microbiota of a European cohort.52 It is interesting to know that allergies and asthma are practically nonexistent in rural African communities.
There is accumulating evidence pointing to an alteration of the gut microbiota in persons with allergies and asthma. 24 Children that live on farms have a lower incidence of asthma than city children.53
Microbiota and metabolismObesity is the result of the increase in the consumption of foods that are high in energy, sugar, and saturated fats. However, it seems that the simple increase in the ingestion of calories does not completely explain the current obesity epidemic. GF mice do not gain weight when they are exposed to high-fat and high-carbohydrate diets, leading to the supposition that diet is not sufficient for inducing obesity.
An «obese-type» human microbiota has been described that is associated with excess weight and metabolic syndrome, with an increase in the Firmicutes/Bacteroidetes ratio.54 The Bifidobacteria and Bacteroides spp. appear to be protectors against the development of obesity.55 Obesity could have a microbial component with probable therapeutic implications.
The colonization of GF mice with normal mouse microbiota produces a dramatic increase in fat in 10-14 days, despite reduced food consumption. The capacity to ferment dietary carbohydrates varies widely among microorganisms and evidence points to a greater efficiency of the gut microbiota of overweight individuals to degrade non-digestible vegetable carbohydrates.23 Turnbaugh et al.23 demonstrated that genetically obese mice (ob/ob) have 50% fewer Bacteroidetes and more Firmicutes than their thin siblings. They proved that the microbiota of the obese mice released more calories during digestion than that of the thin mice. The obesity-causing phenotype may be transmissible: the implantation of the obesogenic gut microbiota in GF mice results in increased adiposity in the receptor mouse.23
When normal weight mice are given a typical high-calorie Western diet for 8 weeks (an accepted mechanism for producing obesity in mice), a marked reduction in Bacteroidetes and a clear rise in Firmicutes is also observed.56 Jumpertz et al. administered diets of varied caloric content to 12 thin human subjects and 9 obese ones and compared the ingested calories with the fecal calories. The modification in the microbiota secondary to diet, with a 20% increase in Firmicutes and the corresponding reduction in Bacteroidetes, was associated with an increase in energy recovery of approximately 150kcal.57
These findings have led to the hypothesis that the microbiota of obese individuals may be more efficient in energy extraction than the microbiota of thin individuals.
It is known that situations occurring around the time of birth increase the risk for developing obesity, diabetes, and cardiovascular disease in the adult stage 58 and the initial colonization could be very important for determining the final composition of the permanent microbiota in adults. 59
The following are some of the many metabolic mechanisms that associate the microbiota with obesity and its related disorders, such as diabetes and fatty liver:
- -
Bacterial fermentation of dietary polysaccharides that cannot be digested by the host, with the consequent production of monosaccharides and SCFA. The SCFA are substrates of the colonocytes and precursors of cholesterol and fatty acids, and they are substrates of gluconeogenesis in the liver, all of which optimizes the exploitation of the energy of the diet.
- -
The SCFA bind to specific intestinal endocrine cell receptors (GRP43 and GRP41) that increase the YY peptide, which delays bowel transit, increasing nutrient absorption60 and increasing the levels of leptin, an orexigenic hormone.61
- -
The microbial regulation of some of the host genes that promote the deposit of lipids in the adipocytes.21
- -
The reduction of the intestinal expression of fasting-induced adipose factor (FIAF) also known as the type IV factor similar to angiopoietin that is a circulating inhibitor of lipoprotein lipase, which favors the fatty acid uptake and the expansion of the adipose tissue. The FIAF can also induce coactivator 1 of the peroxisome proliferator-activated receptor gamma that regulates the expression of the enzymes in charge of fatty acid oxidation.22 In fact, the GF mice that lack the 2 FIAF alleles have the same quantity of body fat as the conventional mice,21 and so it is believed that the FIAF can be a mediator of the microbial regulation of the peripheral fat reserves.62
- -
The obese mice have an increase in the methanogenic archaea, which is associated with a lower partial hydrogen pressure, optimizing the bacterial fermentation velocity.63,64
- -
The hepatic increase in the portal circulation's monosaccharide uptake activates key transcriptional factors, such as ChREBP, that regulate lipogenesis.65
- -
The microbiota increases the vascularization induced by inflammation and the blood flow of the mucosa that, in turn, increases nutrient absorption.66
- -
Gut microbiota is capable of promoting a state of low-grade systemic inflammation, insulin resistance, and of increasing the cardiovascular risk through mechanisms that include exposure to bacterial products, particularly the LPS derived from Gram-negative bacteria. This has been called metabolic endotoxemia.67 Clemente-Postigo et al. recently demonstrated an association between postprandial triglyceride levels and an increase in bacterial endotoxins after a high-fat diet.68 Changes in the gut microbiota, the increase in the intestinal permeability, and endotoxemia possibly play an important role in the development of a low-grade chronic inflammatory state in the host that contributes to the development of obesity and chronic metabolic diseases such as non-alcoholic fatty liver disease (NAFLD).67,69
- -
In recent years, importance has been taken away from BMI as a metabolic syndrome predictor and the concept that visceral fat is responsible for this problem has gained strength. Visceral fat secretes close to 250 proteins,2 such as visceral growth factor, IL-6, the plasminogen activator inhibitor, TNF-α and reactive C protein, all of which are implicated in inflammation.70 This leads to the idea that obesity with its metabolic consequences and accompanying diseases could have an important microbial component, with probable therapeutic implications.
Recent studies begin to profile the association between dysbiosis and gastrointestinal diseases. Important differences have been demonstrated in the microbiota of patients with IBS compared with healthy controls; the Firmicutes/Bacteroidetes relation was shown to be twice as high in the IBS patients (P<0.0002).71 The patients with IBS have fewer Lactobacillus and Bifidobacterium spp. than the healthy controls.72 The bacteria mentioned before bind to epithelial cells and inhibit the adherence of pathogenic bacteria, they do not produce gas upon fermenting the carbohydrates, and they inhibit the Clostridia spp.73 Probiotics modify the colonic fermentation and stabilize the colonic microbiota. Various studies with probiotics have shown an improvement in flatulence and abdominal bloating.74 There are interesting findings in the recent studies on IBS in adults and children. Saulnier et al. found a significantly higher percentage of proteobacteria in children with IBS and they could classify the IBS subtypes based on a limited series of bacteria. Interestingly, a new microbe similar to Ruminococcus was associated with IBS.75
Studies carried out over the past decade identified an association between IBS and the bacterial overpopulation detected through breath tests with the administration of oral lactulose or glucose. In IBS patients, Pimentel et al. demonstrated a 35% symptom improvement upon administering a non-absorbable antibiotic (neomycin) compared with an 11.4% improvement with a placebo. When only the patients in whom the elimination of the bacterial overpopulation was demonstrated after antibiotic use were taken into account, there was improvement in 75% of those patients.76 This line of investigation has been taken with caution due to the difficulties in diagnosing bacterial overpopulation through breath tests. In addition, the use of antibiotics with little absorption, such as neomycin, is not exempt from secondary effects. More recently, 2 phase III studies that were double-blinded and controlled with placebo (TARGET 1 and TARGET 2) were conducted on patients with IBS without constipation and treated with rifaximin, a non-absorbable antibiotic, at a dose of 550mg 3 times a day for 2 weeks, to evaluate IBS symptom improvement. Significantly, more patients in the rifaximin group had a better overall improvement in IBS symptoms, 40.7% vs 31.7%, P=0.01, and improvement in the sensation of abdominal bloating, 40.2% vs 30.3%, P=0.001.77
The studies that show the existence of a gut-brain-microbiota axis are surprising.78 It has been shown that the microbial content of the postnatal gastrointestinal tract in mice is critical for the development of adequate responses to stress in later stages of life. It has also been shown that there is a critical window in the early stages of life in which colonization should occur in order to ensure normal development of the hypothalamic-pituitary-adrenal axis.79
Methane (CH4) is one of the gases present in the human gut and is produced by anaerobic bacterial fermentation. CH4 has been described as being able to affect bowel transit velocity, reduce the secretion of serotonin, and has been associated with IBS, diverticulosis, and colon cancer.80 The main CH4-producing microorganism is Methanobrevibacter smithii, belonging to the Archaea domain.81 Prolonged bowel transit times have been demonstrated in CH4-producing adults.82 A recent study evaluated CH4 production in 629 patients with intestinal symptoms through a glucose breath test and 32.3% of the patients were CH4 producers. The excretion of this gas could be significantly correlated with chronic constipation and it was higher in patients with constipation compared with healthy individuals and much higher than in the patients with diarrhea.83
Crohn's diseaseMany studies have suggested the presence of dysbiosis in the intestine of patients with Crohn's disease, compared with healthy individuals.84 Healthy twins tend to have a very similar microbiota, but when one of the twins has Crohn's disease, the intestinal composition changes greatly, especially in patients with ileal inflammation.85
Celiac diseaseA marker of active celiac disease is the production of cytokines by intestinal T lymphocytes in individuals that are carriers of certain class II MHC alleles. It has been suggested that dysbiosis is another risk factor for celiac disease. In fact, a «Swedish celiac disease epidemic» was described86 and bacterial candidates have been isolated as etiologic factors that were later able to be isolated in patients born during the epidemic. Dysbiosis and the bacteria associated with celiac disease can be a risk factor for the development of the disease, whether it is by direct influence in the immune responses of the mucosa or upon increasing the inflammatory response to gluten.87
Non-alcoholic steatohepatitis/non-alcoholic fatty liver diseaseUpon conventionalizing GF mice from the cecum of normal mice, Backhed et al. demonstrated an increase in fat in the liver.21 NASH and NAFLD have been associated with bacterial overpopulation and increased intestinal permeability, even though not all studies are concordant.62
Various bacterial products can be potentially hepatotoxic: phenols, ammonium, ethanol, and others.88 An increase in ethanol production has been described in obese patients.89 It is thought that the main bacterial product implicated in NASH and NAFLD is LPS, the active component of the endotoxins of the bacterial wall, released through bacterial death in the intestine. LPS goes through capillary translocation by means of a TLR-4-dependent mechanism and is absorbed together with dietary lipids.90 LPS absorption in turn activates TNF-α, IL-1, and IL-6 production. The signals that TLR-4 awakens promote insulin resistance, hepatic steatosis, inflammation, and fibrogenesis.88
Chronic infusion of LPS at low doses in mice causes obesity and an increase in body fat percentage, insulin resistance, macrophage infiltration into the adipose tissue, and hepatic steatosis.91 Studies in humans have also shown that endotoxemia is a risk factor for the development of NASH/NAFLD. Two studies on patients with NAFLD diagnosed through biopsy showed an increase in endotoxemia when compared with healthy individuals.92,93
In conclusion, modern analysis of the bacterial genome is of great interest and has opened a field of investigation that can explain the close relationship between the microbiome and humans, and can help answer the questions about modern «epidemics»: the autoimmune, allergic, and metabolic diseases; but it especially offers us the possibility of attempting to revert them through the manipulation of the components of the microbiota. Evidence shows that the microbiota is stable over time and that some effects of early-stage human colonization are irreversible. And so these questions arise: Do we have the capacity to prevent alterations in the microbiota that are due to an excess of hygiene and the lack of contact with healthy microorganisms? Can we manipulate the microbiota of an individual in a permanent or at least long-term manner?
Financial disclosureNo financial support was received in relation to this article.
Conflict of interestThe authors declare that there is no conflict of interest.
Please cite this article as: Icaza-Chávez ME. Microbiota intestinal en la salud y la enfermedad. Revista de Gastroenterología de México. 2013;78:240–248.